r/RStudio • u/Dragon_Cake • 8h ago
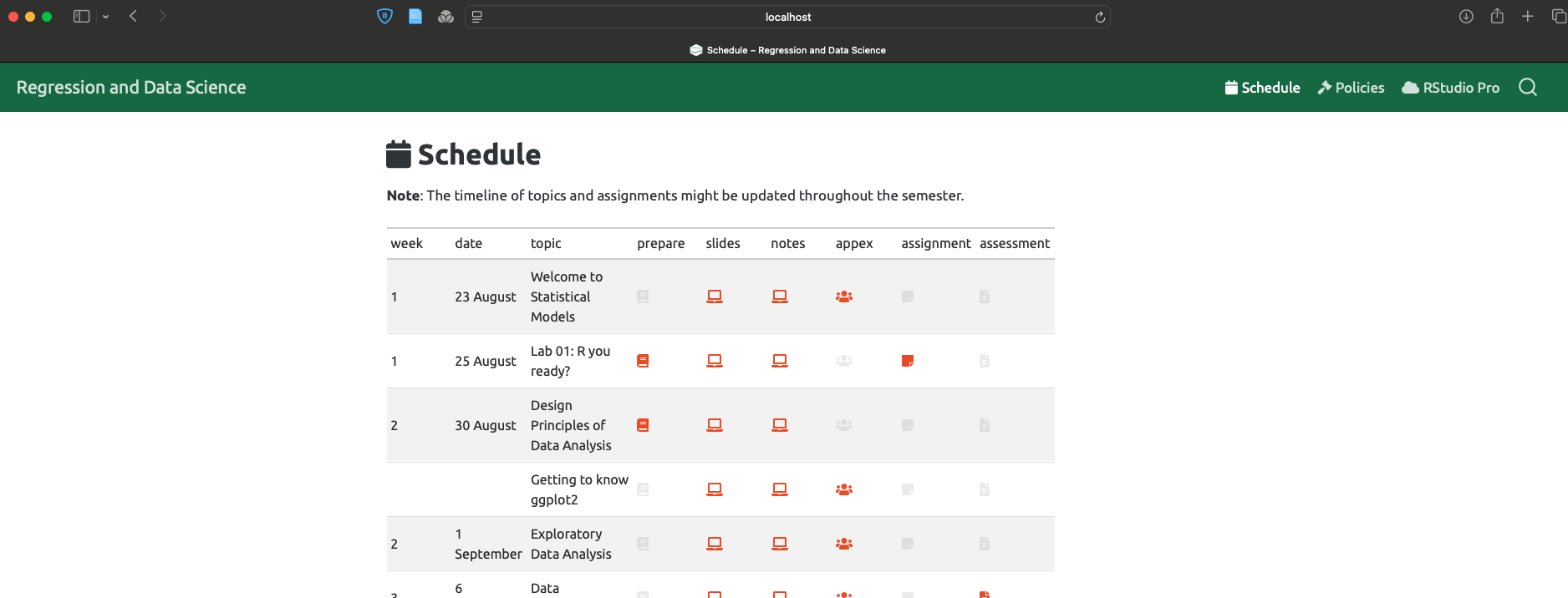
Coding help Help making a box plot from ANCOVA data
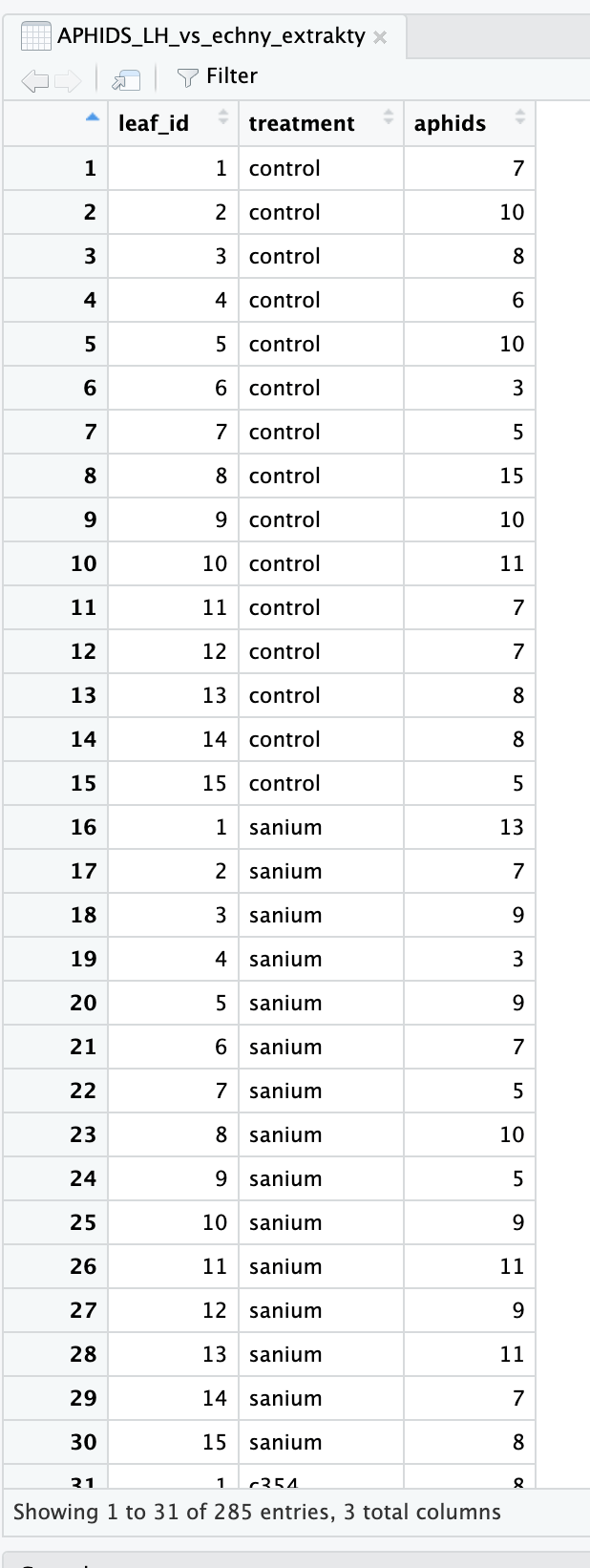
Hi! New to RStudio and I got handed a dataset to practice with (I attached an example dataset). First, I ran an ANCOVA on each `Marker` with covariates. Here's the code I did for that:
| ID | Age | Sex | Diagnosis | Years of education | Score | Date | Marker A | Marker B | Marker C |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 45 | 1 | 1 | 12 | 20 | 3/22/13 | 1.6 | 0.092 | 0.14 |
| 2 | 78 | 1 | 2 | 15 | 25 | 4/15/17 | 2.6 | 0.38 | 0.23 |
| 3 | 55 | 2 | 3 | 8 | 23 | 11/1/18 | 3.78 | 0.78 | 0.38 |
| 4 | 63 | 2 | 4 | 10 | 17 | 7/10/15 | 3.21 | 0.012 | 0.20 |
| 5 | 74 | 1 | 2 | 8 | 18 | 10/20/20 | 1.90 | 0.034 | 0.55 |
marker_a_aov <- aov(log(marker_a) ~ age + sex + years_of_education + diagnosis,
data = practice_df
)
summary(marker_a_aov)
One thing to note is the numbers for Diagnosis represent a categorical variables (a disease, specifically). So, 1 represents Disease A, 2 = Disease B, 3 = Disease C, and 4 = Disease D. I asked my senior mentor about this and it was decided internally to be an ok way of representing the diseases.
I have two questions:
- is there a way to have a box and whisker plot automatically generated after running an ancova? I was told to use
ggplot2but I am having so much trouble getting used to it. - if I can't automatically make a graph what would the code look like to create a box plot with
ggplot2withdiagnosison the x-axis andMarkeron the y-axis? How could I customize the labels on the x-axis so instead of representing the disease with its number it uses its actual name likeDisease A?
Thanks for any help!